Articles
- Page Path
- HOME > J Mov Disord > Volume 10(1); 2017 > Article
-
Review Article
MicroRNA Biomarkers in Neurodegenerative Diseases and Emerging NanoSensors Technology - Pratik Shah1,2, Seok Keun Cho3, Peter Waaben Thulstrup4, Morten Jannik Bjerrum4, Phil Hyu Lee5, Ju-Hee Kang6, Yong-Joo Bhang7, Seong Wook Yang1,3
-
Journal of Movement Disorders 2017;10(1):18-28.
DOI: https://doi.org/10.14802/jmd.16037
Published online: January 18, 2017
1UNIK Center for Synthetic Biology, University of Copenhagen, Copenhagen, Denmark
2Department of Biomedical Engineering, University of California Irvine, Irvine, CA, USA
3Department of Systems Biology, College of Life Science and Biotechnology, Yonsei University, Seoul, Korea
4Department of Chemistry, University of Copenhagen, Copenhagen, Denmark
5Department of Neurology, Severance Biomedical Science Institute, Yonsei University College of Medicine, Seoul, Korea
6Department of Pharmacology, Hypoxia-related Disease Research Center, Inha University School of Medicine, Incheon, Korea
7Seoulin Bioscience Co., Ltd., Seongnam, Korea
- Corresponding author: Pratik Shah, PhD, Department of Biomedical Engineering, University of California Irvine, Irvine 92697, CA, USA Tel: +1-949-501-6270 E-mail: pshah2@uci.edu
Copyright © 2017 The Korean Movement Disorder Society
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
ABSTRACT
- MicroRNAs (miRNAs) are essential small RNA molecules (20–24 nt) that negatively regulate the expression of target genes at the post-transcriptional level. Due to their roles in a variety of biological processes, the aberrant expression profiles of miRNAs have been identified as biomarkers for many diseases, such as cancer, diabetes, cardiovascular disease and neurodegenerative diseases. In order to precisely, rapidly and economically monitor the expression of miRNAs, many cutting-edge nanotechnologies have been developed. One of the nanotechnologies, based on DNA encapsulated silver nanoclusters (DNA/AgNCs), has increasingly been adopted to create nanoscale bio-sensing systems due to its attractive optical properties, such as brightness, tuneable emission wavelengths and photostability. Using the DNA/AgNCs sensor methods, the presence of miRNAs can be detected simply by monitoring the fluorescence alteration of DNA/AgNCs sensors. We introduce these DNA/ AgNCs sensor methods and discuss their possible applications for detecting miRNA biomarkers in neurodegenerative diseases.
- As the fundamental unit of living organisms, the cell runs thousands of biochemical reactions with high accuracy, precision and efficiency. The systemic regulation of cellular processes involves a variety of regulatory components and pathways. To study these sophisticated systems, researchers rely on cellular biomarkers as the representatives of any given cellular reaction. The biomarkers provide key information about the normal biological processes, as well as the pathogenic or disease conditions that arise in response to external signals or internal imbalances. One highly attractive class of biomarkers comprises the nucleic acids (NAs), i.e., DNA and RNA, because of their easy extraction from non-invasive samples such as blood, urine and stool for prognosis and diagnosis [1,2]. One of the most widely used biomarkers, with a huge potential for practical applications, is a class of small RNA molecules known as microRNAs (miRNAs) [3]. miRNAs are short non-coding RNAs that are 21−22 nucleotides in length and are involved in the regulation of gene expression at the post-transcriptional level [4]. Detailed studies have revealed the spatial and temporal occurrence of many miRNAs, as well as their unique biological roles in messenger RNA (mRNA) cleavage and translational suppression. More than 2000 miRNAs have been identified in humans, and many of them are functionally involved in diseases, showing great potential as biomarkers [4-6]. However, for the practical applications of miRNAs as biomarkers, highly reliable, economic, and convenient detection methods are required in addition to the currently available methods. Thus, there has been a surge in the interest of developing improved miRNA detection methods, especially using nanotechnological tools [5-9]. One such advanced tool that has drawn particular attention is the use of the fluorescent properties of silver nanoclusters (AgNCs) [10-12]. When clustered silver atoms approach a size less than 2 nm, due to quantum confinement, AgNCs exhibit unique properties such as strong and stable fluorescence [12,13]. The synthesis of AgNCs in aqueous solutions, using stabilizing scaffolds such as NAs, has allowed for a large number of applications for the detection of biomolecules [14-16]. The fundamental understanding of the role and properties of NAs, as well as their influence on the fluorescent synthesis of AgNCs, has increased in recent years [17-21]. By exploiting such new technology, research groups have developed several reliable methods for NA/AgNCs-based miRNA detection [10,22]. In this review, we discuss the current findings of miRNAs as biomarkers for human diseases, especially neurodegenerative diseases, and how miRNAs can be efficiently determined, with an emphasis on novel nanosensor methods.
INTRODUCTION
- The biological impacts of the small RNA molecules remained unnoticed until the last part of the 20th century, but the discovery of miRNA by the group of Victor Ambros opened up a whole new avenue of research [4]. The miRNAs turned out to be very important regulators of gene expression [4]. Recent studies on their origin, biogenesis and mechanisms of action have greatly advanced our knowledge of the molecular biology of miRNAs [4]. miRNAs are generated through two step-wise processes involving the coordinated functions of a multi-protein complex known as the microprocessor [23-25]. Primary miRNAs are transcribed from MIRNA genes and then processed into precursor miRNAs and further to mature miRNAs by the microprocessor [23]. Mature miRNAs are then loaded onto the so-called RNA-Induced Silencing Complex to recognize target mRNAs through sequence complementarity. The complex of the protein machinery with a loaded miRNA can either remove target mRNA or repress the translation of mRNA [26,27]. The levels of miRNAs are often differentially modulated by the growth and developmental stage of organisms, specific cell and tissue types, and environmental stimuli [3,28-33].
BIOLOGY OF MICRORNAS
- Many studies have suggested that miRNAs can be important biomarkers in a variety of diseases, including cancer, diabetes, cardiovascular disease, aging, asthma, autoimmune disease, kidney diseases, and neurodegenerative diseases [3,34-46]. Due to the potential of miRNA as biomarkers, several studies have predicted that monitoring methods for miRNAs will be extensively useful for curing those diseases in the near future [42,47-50]. Among the defined miRNAs, we focused our discussion on miRNAs associated with neurodegenerative diseases in this review. Neurodegenerative disorders−Alzheimer’s disease (AD), Huntington’s disease (HD), Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS), and prion diseases− are defined by the progressive loss of specific neuronal cells in the central nervous system. These diseases commonly cause significant defects in motor and cognitive ability. Recently, a number of studies have revealed that specific miRNAs are differentially expressed in the human brain and, more importantly, some of the miRNAs modulate genes associated with specific neurodegenerative disorders. For instance, Harraz et al. [51] showed that among 224 miRNAs sequenced, miR-133b, miR218-2, miR-15b, miR101-1, miR107, miR-335, and miR-345 were notably down-regulated in PD patients. miR-133b seems to be functional as a negative regulator of Pitx3, an important transcriptional factor that is involved in the differentiation of dopaminergic neurons [52-54]. Interestingly, miR-133 and Pitx3 modulate the expression of each other through a negative feedback regulation. Pitx3 induces MIR-133 gene expression, which in turn targets Pitx3 to be degraded [55]. The profiling of miRNAs in PD brains uncovered decreased expression of miR-34b and miR-34c in the affected brain areas [56]. The reduction of miR-34b/34c inversely enhanced the expression of α-synuclein in human dopaminergic SH-SY5Y cells [57]. The sequence variations in the miR-433 recognition site of fibroblast growth factor 20 (FGF20) may disrupt the miR-433-mediated silencing of FGF20, and that results in the increased expression of α-synuclein, which confers risk for PD [58]. Three differentially regulated miRNAs−miR-1, miR-22*, and miR-29−were identified in the blood samples of PD patients compared to healthy subjects, showing the feasibility of miRNAs as biomarkers [59].
- β-amyloid cleavage enzyme 1 (BACE-1) is required for the cleavage of amyloid precursor protein (APP), which thereby generates toxic Aβ species [60]. The expression of BACE-1/β-secretase is inversely correlated with the levels of miRNAs from the miR-29a/b-1 family, showing that BACE-1/β-secretase is a target of this family miRNA [61]. Interestingly, in a group of AD patents, the expression of miR-29a/b-1 clusters was significantly down-regulated [61,62]. Two additional miRNAs, miR-298 and miR-328, also seem to play essential roles in down-regulating BACE-1/β-secretase expression [61,63]. The level of miR-107 is reduced in the brains of AD patients and that is inversely correlated with the increased levels of BACE-1/β-secretase and cofilin, showing the role of miR-107 in human AD [64,65]. Cyclin-dependent kinase 5, a tau kinase which is dysregulated in AD, and the metalloproteinase ADAM10, which is important for APP processing, are also targeted by miR-107 [66]. miR-15 targets a pathological hallmark gene of AD, extracellular signal-regulated kinase 1, a tau kinase. The level of miR-15 is also decreased in AD patiens [67]. Other studies have demonstrated that the miR-29 family seems to play a role in the modulation of microglial activity. The miR-29 family also targets the microglial modulators insulin-like growth factor-1 (IGF-1) and fractalkine ligand (CX3CL1). Indeed, the increased level of miR-29b was inversely correlated with the expression of IGF-1 and CX3CL1 in human cortical tissue, implying the role of the miR-29 family in brain aging [68]. p53 is highly upregulated in AD and induces tau hyperphosphorylation. The miR-34 family is transcriptionally induced by p53 and is also considered a critical mediator of p53 functions. Accordingly, miR-34 is found at high levels in the hippocampus of AD patients, and the down-regulation of miR-34 rescues some cognitive defects [69,70]. miR-106a and miR-106b are complementary to the transcript of APP, which is increased in AD patients. Consistently, these miRNAs are reduced in the temporal lobe of AD patients. The expression of the transporter ABCA1 is also regulated by miR-106a and miR-106b, confirming the roles of the miR-106 family in AD processes [71]. An miRNA dysfunction in AD could be a cause, but in some cases, it could also be a result of AD progress. For example, miR-181c is down-regulated by Aβ in in vitro hippocampal cultures. This reduction of miR-181c is recapitulated in Aβ-depositing APP23 transgenic mice and in human AD tissue [72]. In contrast, miR-9, miR-125b, and miR-128 are increased in the hippocampal region of AD-affected brains [73]. In the case of miR-146a, it is upregulated in the temporal cortex of AD patients, and complement factor H is a target of miR-146a, indicating that miR-146a mediates AD-related inflammation [74]. Other miRNAs have been associated with AD, such as miR-124, miR-132, and miR-153. Most of them seem to be implicated in APP processing, neuro-inflammation, tau hyper-phosphorylation, and ApoE-lipidization [66,75].
- The pathogenic poly-glutamine expanded huntingtin (HTT) causes HD, a hereditary neurodegenerative disorder. In normal neurons, HTT forms a complex with repressor element 1 silencing transcription factor (REST) in the cytoplasm, which suppresses the translocation of REST into the nucleus. The REST-HTT interaction can be abolished by the pathogenic modification of HTT, which consequently leads to neuronal death [76,77]. The transcript of REST is targeted by miR-9; therefore, the reduction of miR-9 in HD brains could cause the accumulation of free REST protein. Indeed, the cerebral cortex of HD brains have considerably reduced levels of miR-9, showing the importance of miR-9 in HD [78]. miR-132 is notably down-regulated in the post-mortem brains of HD patients. The reduction of miR-132 is known to increase its target, p250GAP, encoding a member of the group of GTPase-activating proteins, which inhibit neurite outgrowth, resulting in HD progress [79,80].
- Although we have hitherto discussed miRNAs dysregulation in AD, PD, and HD, there are many miRNAs that are directly or indirectly involved in other neurological disabilities (Table 1), such as epilepsy, ALS, traumatic brain injury, and prion diseases. Recent studies have shed light on the impacts of numerous miRNAs on the pathogenesis and progression of neurodegenerative diseases. However, comprehensive profiles of the effects of different sets of miRNAs that are related to a specific disease still remain elusive. Not only for disease diagnosis but also for prognosis, the expression profile of miRNAs must be precisely and efficiently characterized. To meet the demand, a variety of methods for miRNA profiling have been developed, some of which are discussed below.
MIRNA BIOMARKERS IN HUMAN NEURODEGENERATIVE DISEASES
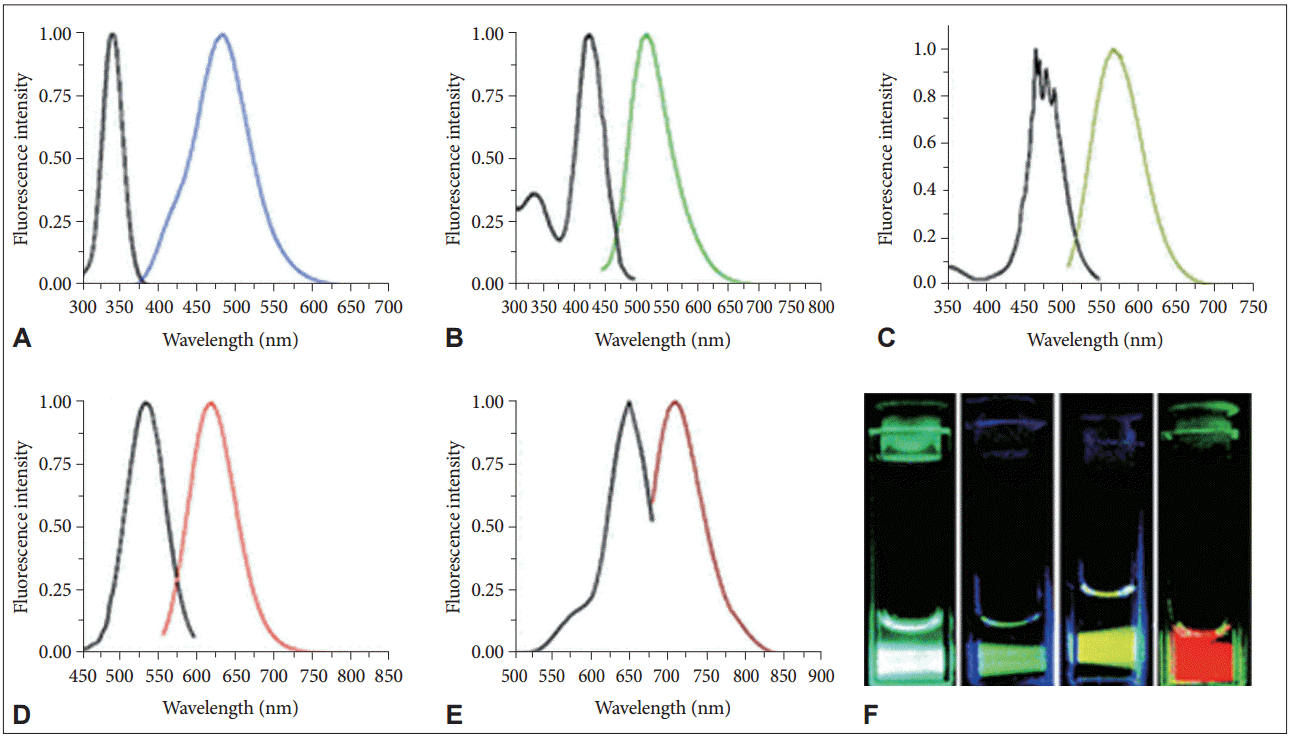
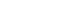
- Metal nanoclusters (MNCs) exhibit size-dependent distinct optical properties unlike their bulk counterpart material [12,13,81]. When the metal size approaches the Fermi wavelength, the continuous density of states becomes discrete and all the electronic wave-functions are overlapped [82]. In this size regime, MNCs may behave as molecular systems, where energy levels are separated to allow direct electronic transitions due to quantum confinement (Figure 1). Because of their behavior as “pseudo-atoms,” the metal clusters exhibit unique properties such as strong photoluminescence [83,84]. Among MNCs, AgNCs have gained more prominence due to their brighter fluorescence emission. AgNCs are unstable in solution, and they readily aggregate to form large particles or aggregates lacking any fluorescence properties. For biological application purposes, a water soluble and biocompatible encapsulating ligand is required to restrict the growth of AgNCs in the specific size regime (< 2 nm) [11,12]. In recent years, many studies have shown the development of AgNCs using different stabilizing templates, such as dendrimers, peptides, and thiolates [81,85-88]. Among the various stabilizing templates for emissive AgNCs formation, we here focus on a type of biocompatible template, NAs, for generating AgNCs. Due to the biological implications of DNA and RNA, NAs have attracted great attention as scaffold templates for emissive AgNCs. The first major advancement in this direction was achieved by Dickson’s group [11]. Not only could a strong emission be measured from the DNA AgNCs, but as shown by Dickson’s group, the emission wavelength of the AgNCs could also be altered (Figure 2) [89,90]. Furthermore, the structure and sequences of NAs can create an excellent interface between the fluorescent properties of AgNCs and biological applications. Therefore, a whole new avenue of applications was opened with the findings from Dickson’s group, proving the potential of NAs as a stabilizing template for AgNCs [11]. In subsequent studies, many research groups focused on understanding the fundamental mechanisms underlying the utility of AgNCs as fluorophores. Although these studies suggested many interesting findings on the nature of AgNCs that are worthwhile to discuss in detail, we here focus on discussing the applications of DNA/AgNCs sensors for miRNAs.
INTRODUCTION TO SILVER NANOCLUSTERS
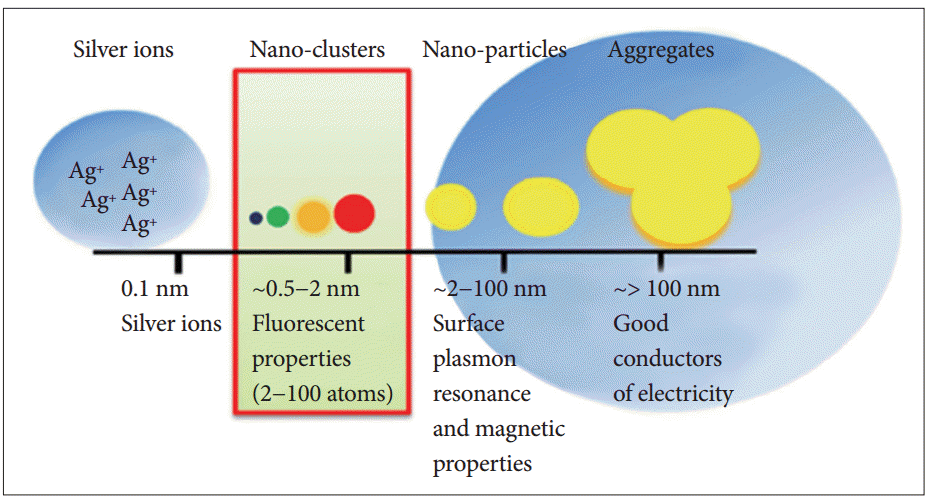
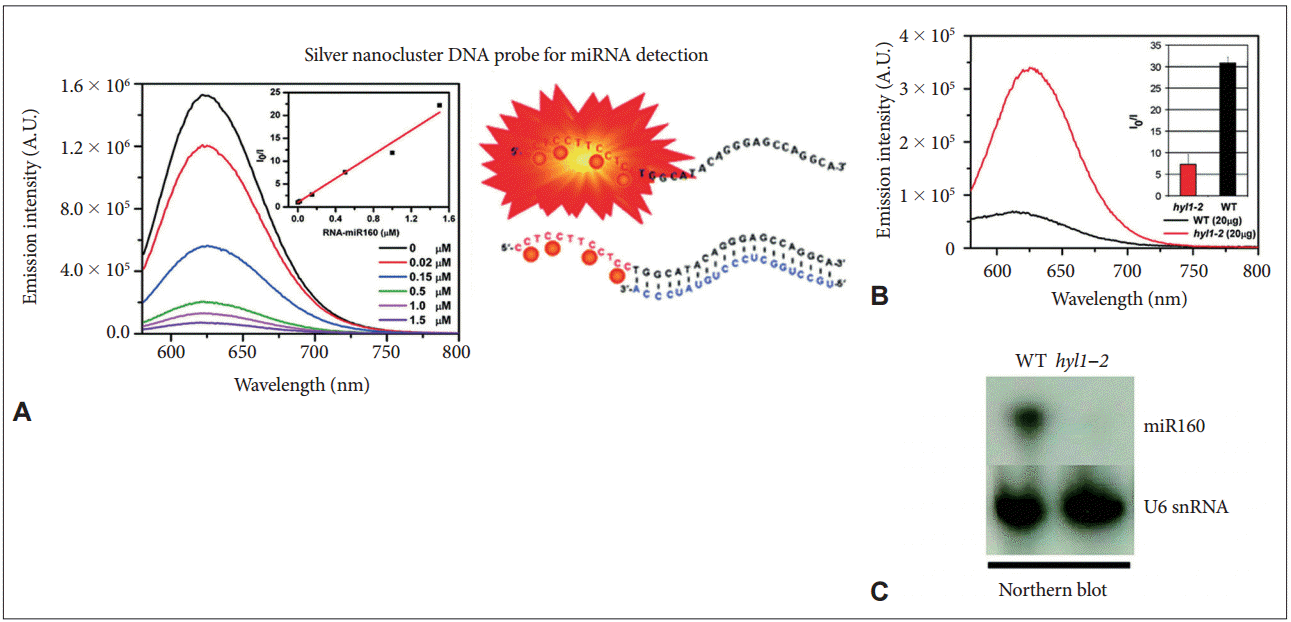
- For the first time, Petty et al. [91] demonstrated the potential value of DNA/AgNCs for the detection of DNA, where a bi-functional DNA sensor was used. One component of the sensor was composed of a cytosine-rich sequence, and the other part was a sequence complementary to the target DNA sequences. The presence of the target DNA sequence enhanced the fluorescence of the DNA/AgNCs sensor approximately 2−3-fold, enabling the sensor to be used in a turn-on method for detecting a specific DNA fragment. In a further development of the system, the same group showed that the wavelength shift, rather than altered fluorescence intensity, can be used to detect a target DNA fragment [91]. By applying the potential of DNA/AgNCs-based methods, Yang and Vosch [10] reported for the first time a DNA/AgNCs-based method for miRNA detection. A DNA-sensor consists of two components, a cytosinerich scaffold and a DNA sequence complementary to the target miRNA. The DNA sensor generates a bright red fluorescence within 1 hour and drops the fluorescence in the presence of target miRNA. This “turn-off”-based method allows the quantitative detection of miRNA down to a picomole level (Figure 3). Because of the huge diversity of miRNA sequences in Arabidopsis and humans, Shah et al. [20] attempted to systemically design DNA/AgNCs sensors by changing only the target miRNA sensing sequences. However, they found that the systemic designing of DNA/AgNCs sensors was not simply established by replacing the target sensing sequences for each miRNA. Through detailed analysis, they suggested that the secondary structure of DNA/AgNCs sensors, including the silver encapsulating C-rich scaffold, is essential for the generation of fluorescence and target recognition [20]. Based on this finding, they reconstituted the C-rich scaffold and target recognition sequences of the non-functional DNA/AgNCs sensor (for miR172 in plants) to be structured, and that was eventually functional in detecting target miRNA. Next, Shah et al. [92] demonstrated that RNA and a DNA/RNA chimera can be an efficient sensor for a specific group of human miRNAs because of the rotational freedom of RNA backbones. By introducing RNA backbones, they further showed that nonfunctional DNA/AgNCs sensors (for let-7a and miR- 200 in humans) can be successfully converted into functional RNA/DNA-chimera/AgNCs sensors [93]. The newly developed chimera sensors were highly sensitive to recognize let-7a and miR-200 in human cell lines. Furthermore, to improve the designing system for DNA/AgNCs sensors, Shah et al. [94] suggested a locking-to-unlocking system by which several DNA/AgNCs sensors were successfully constructed for miR-21, miR-18a, and miR-27b (Figure 4).
- These studies also demonstrated the pragmatic potential of DNA/AgNCs-based sensors by using biological samples. For instance, the levels of target miR160 in wild-type and in hyl1-2 mutant of Arabidopsis were determined with a DNA/AgNCs sensor [10]. Let-7a is now used as a biomarker for many cancers in humans, such as colorectal cancer and breast cancer. By using chimera/AgNCs sensors, the expression of let-7a was successfully observed in HT-29 (a colorectal adenocarcinoma) and MDA-MB-231 (a breast cancer cell line); miR-200c was monitored only in the HT-29 cell line [93]. Likewise, miR-21 and miR-27b are considered as specific biomarkers of cancers such as MCF-7 (a breast cancer cell line) and PANC-1 (a non-endocrine pancreatic cancer cell line), respectively. The differential expression of miR-21 and miR-27b in the cell lines was successfully monitored by the locking-to-unlocking DNA/AgNCs methods [94]. Yang’s group further applied the method to study the processing pathway of miRNA in plants and identified that the deficiency of CONSTITUTIVE PHOTOMORPHOGENETIC 1, a negative regulator of photomorphogenesis, led to a dramatic reduction of miRNAs [95]. The detection of miRNA levels by the DNA/AgNCs-based method allowed them to rapidly screen out a novel component in plant miRNA biogenesis.
- In addition to these methods, Zhang et al. [96] suggested the use of target-assisted isothermal exponential amplification coupled with fluorescent DNA/AgNCs. By using two enzymes, DNA polymerase and nicking endonuclease, the polymerization of a designed DNA sensor can be initiated upon the binding of target miRNA. They presented that miR-21 and miR-141 could be detected with a high specificity and sensitivity, and claimed that the sensitivity was similar to the routinely used quantitative real-time reverse transcription PCR (qRT-PCR)-based method. The outcome of this method was verified by qRT-PCR in parallel. Xia et al. [97] suggested a new type of AgNCs/hairpin DNA sensor where a 5-TCC/CCC-3’-overhang for embedding emissive AgNCs was attached to a target sensing DNA sequence. The fluorescence of the sensor was diminished in the presence of the target RNA sequence with complementarity to the sensing DNA sequence. Two hairpin DNA-templated AgNCs (AgNCs/HpDNA) sensors were combined with a strand-displacement amplification (SDA) procedure to detect miR-16-5p and miR-19b-3p, which are known as biomarkers of gastric cancer. This method demonstrated that the effect of G-rich fluorescence enhancement can be successfully implemented in the SDA reaction for the rapid and specific detection of miRNAs. Dong et al. [98] demonstrated that a conformational molecular beacon coupled with target recycling amplification can be a facile and sensitive miRNA biosensor. In the method, the competition displacing between target miRNA and Hg2+ provides the biosensor with high sequence specificity. Although the functionality of a DNA/AgNCs-based method has been proven to date with only miRNA biomarkers in cancer cells and tissue samples from cancer patients, and because some of them are also functional in neurodegenerative diseases, the potential of the method is evidently promising for detecting miRNA biomarkers in neurodegenerative diseases.
DNA/AGNCS-BASED METHODS FOR MIRNA DETECTION
- Each of the conventional methods for miRNA detection, such as small RNA blotting, qRT-PCR, and microarrays, has its own individual drawbacks. For instance, small RNA blotting is labor intensive and requires radioactive-labeling or very complicated DIG-labeling. In general, small RNA blot analysis takes at least 2 days to evaluate the levels of miRNAs in samples, and its sensitivity is relatively lower than other methods. However, this method has a major advantage that reflects the actual levels of miRNAs in samples, as it reads the signals from direct hybridization between a probe and target miRNA. qRT-PCR and microarrays are costly in terms of operational materials and equipment and have nonspecific readout problems. The qRT-PCR-based method detects target miRNAs by PCR, and it extensively relies on the success of cDNA synthesis, which is initiated by priming miRNAs with a short adaptor. Failure of the primers to conjugate with the miRNAs could result in false negatives. In the case of microarray analysis, a probe must hybridize with the miRNA in a fixed temperature, even though miRNAs vary in their melting temperatures. This intrinsic limitation often causes readouts to be false positives or false negatives [99]. Therefore, many research groups have extensively investigated ways to build efficacious tools for rapid, simple and specific miRNA detection as alternatives to the conventional methods. To date, many new methods have been suggested, such as nanomaterial-derived methods, nonPCR-based amplification methods, electrochemical methods, etc. The newly developed methods focus on simplicity, lower cost, and better sensitivity and specificity as the common concepts. Here, we have discussed a class of the miRNA detection methods by exploiting the photoluminescent characteristics of AgNCs. Compared to other methods, the AgNCs-based methods have many advantages in monitoring miRNAs, such as the ease of applicability, the extremely low cost of materials, a wide range of fluorescence properties, and the feasibility of both on-and-off signal platforms. Therefore, we suggest that by applying DNA/AgNCs sensor methods, miRNAs biomarkers and their expression profiles in neurodegenerative diseases could be easily investigated without cost or labor problems.
DISCUSSION
- This research was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) and funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI16C1118).
Acknowledgments


- 1. Cappelletti V, Appierto V, Tiberio P, Fina E, Callari M, Daidone MG. Circulating biomarkers for prediction of treatment response. J Natl Cancer Inst Monogr 2015;2015:60–63.ArticlePubMedPDF
- 2. Madic J, Kiialainen A, Bidard FC, Birzele F, Ramey G, Leroy Q, et al. Circulating tumor DNA and circulating tumor cells in metastatic triple negative breast cancer patients. Int J Cancer 2015;136:2158–2165.ArticlePubMed
- 3. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857–866.ArticlePubMedPDF
- 4. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843–854.ArticlePubMed
- 5. Cissell KA, Rahimi Y, Shrestha S, Hunt EA, Deo SK. Bioluminescence-based detection of microRNA, miR21 in breast cancer cells. Anal Chem 2008;80:2319–2325.ArticlePubMed
- 6. Liu CG, Calin GA, Volinia S, Croce CM. MicroRNA expression profiling using microarrays. Nat Protoc 2008;3:563–578.ArticlePubMedPDF
- 7. Várallyay E, Burgyán J, Havelda Z. MicroRNA detection by northern blotting using locked nucleic acid probes. Nat Protoc 2008;3:190–196.ArticlePubMedPDF
- 8. Fang S, Lee HJ, Wark AW, Corn RM. Attomole microarray detection of microRNAs by nanoparticle-amplified SPR imaging measurements of surface polyadenylation reactions. J Am Chem Soc 2006;128:14044–14046.ArticlePubMedPMC
- 9. Gao Z, Yang Z. Detection of microRNAs using electrocatalytic nanoparticle tags. Anal Chem 2006;78:1470–1477.ArticlePubMed
- 10. Yang SW, Vosch T. Rapid detection of microRNA by a silver nanocluster DNA probe. Anal Chem 2011;83:6935–6939.ArticlePubMed
- 11. Petty JT, Zheng J, Hud NV, Dickson RM. DNA-templated Ag nanocluster formation. J Am Chem Soc 2004;126:5207–5212.ArticlePubMed
- 12. Zheng J, Nicovich PR, Dickson RM. Highly fluorescent noble-metal quantum dots. Annu Rev Phys Chem 2007;58:409–431.ArticlePubMedPMC
- 13. Pettibone JM, Gigault J, Hackley VA. Discriminating the states of matter in metallic nanoparticle transformations: what are we missing? ACS Nano 2013;7:2491–2499.ArticlePubMed
- 14. Huang Z, Pu F, Lin Y, Ren J, Qu X. Modulating DNA-templated silver nanoclusters for fluorescence turn-on detection of thiol compounds. Chem Commun (Camb) 2011;47:3487–3489.ArticlePubMed
- 15. Park J, Lee J, Ban C, Kim WJ. An approach toward SNP detection by modulating the fluorescence of DNA-templated silver nanoclusters. Biosens Bioelectron 2013;43:419–424.ArticlePubMed
- 16. Wang X, Lin R, Xu Z, Huang H, Li L, Liu F, et al. N-acetylcysteine induced quenching of red fluorescent oligonucleotide-stabilized silver nanoclusters and the application in pharmaceutical detection. Anal Chim Acta 2013;793:79–85.ArticlePubMed
- 17. Gwinn EG, O’Neill P, Guerrero AJ, Bouwmeester D, Fygenson DK. Sequence-dependent fluorescence of DNA-hosted silver nanoclusters. Adv Mater 2008;20:279–283.Article
- 18. Sengupta B, Ritchie CM, Buckman JG, Johnsen KR, Goodwin PM, Petty JT. Base-directed formation of fluorescent silver clusters. J Phys Chem C 2008;112:18776–18782.Article
- 19. Yeh HC, Sharma J, Han JJ, Martinez JS, Werner JH. A DNA--silver nanocluster probe that fluoresces upon hybridization. Nano Lett 2010;10:3106–3110.ArticlePubMed
- 20. Shah P, Rørvig-Lund A, Chaabane SB, Thulstrup PW, Kjaergaard HG, Fron E, et al. Design aspects of bright red emissive silver nanoclusters/DNA probes for microRNA detection. ACS Nano 2012;6:8803–8814.ArticlePubMed
- 21. Schultz D, Gardner K, Oemrawsingh SS, Markešević N, Olsson K, Debord M, et al. Evidence for rod-shaped DNA-stabilized silver nanocluster emitters. Adv Mater 2013;25:2797–2803.ArticlePubMed
- 22. Liu YQ, Zhang M, Yin BC, Ye BC. Attomolar ultrasensitive microRNA detection by DNA-scaffolded silver-nanocluster probe based on isothermal amplification. Anal Chem 2012;84:5165–5169.ArticlePubMed
- 23. Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 2002;21:4663–4670.ArticlePubMedPMC
- 24. Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev 2002;16:1616–1626.ArticlePubMedPMC
- 25. Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004;432:235–240.ArticlePubMed
- 26. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 2008;9:102–114.ArticlePubMedPDF
- 27. Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 2011;12:99–110.ArticlePubMedPDF
- 28. Aumiller V, Förstemann K. Roles of microRNAs beyond development--metabolism and neural plasticity. Biochim Biophys Acta 2008;1779:692–696.ArticlePubMed
- 29. Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol 2008;9:839–845.ArticlePubMedPDF
- 30. Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. MicroRNAs in cancer management. Lancet Oncol 2012;13:e249–e258.ArticlePubMed
- 31. Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004;16:2001–2019.ArticlePubMedPMC
- 32. Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK. A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 2005;15:2038–2043.ArticlePubMed
- 33. Zhao B, Liang R, Ge L, Li W, Xiao H, Lin H, et al. Identification of drought-induced microRNAs in rice. Biochem Biophys Res Commun 2007;354:585–590.ArticlePubMed
- 34. Kato M, Castro NE, Natarajan R. MicroRNAs: potential mediators and biomarkers of diabetic complications. Free Radic Biol Med 2013;64:85–94.ArticlePubMedPMC
- 35. Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by microRNAs. Nat Rev Immunol 2008;8:120–130.ArticlePubMedPDF
- 36. Beckman JA, Creager MA, Libby P. Diabetes and atherosclerosis: epidemiology, pathophysiology, and management. JAMA 2002;287:2570–2581.ArticlePubMed
- 37. Ji X, Takahashi R, Hiura Y, Hirokawa G, Fukushima Y, Iwai N. Plasma miR-208 as a biomarker of myocardial injury. Clin Chem 2009;55:1944–1949.ArticlePubMed
- 38. Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C, et al. Circulating microRNAs in patients with coronary artery disease. Circ Res 2010;107:677–684.ArticlePubMed
- 39. Neal CS, Michael MZ, Pimlott LK, Yong TY, Li JY, Gleadle JM. Circulating microRNA expression is reduced in chronic kidney disease. Nephrol Dial Transplant 2011;26:3794–3802.ArticlePubMedPDF
- 40. Zhao C, Dong J, Jiang T, Shi Z, Yu B, Zhu Y, et al. Early second-trimester serum miRNA profiling predicts gestational diabetes mellitus. PLoS One 2011;6:e23925. ArticlePubMedPMC
- 41. Natarajan R, Putta S, Kato M. MicroRNAs and diabetic complications. J Cardiovasc Transl Res 2012;5:413–422.ArticlePubMedPMC
- 42. Tijsen AJ, Pinto YM, Creemers EE. Circulating microRNAs as diagnostic biomarkers for cardiovascular diseases. Am J Physiol Heart Circ Physiol 2012;303:H1085–H1095.ArticlePubMed
- 43. Grasso M, Piscopo P, Confaloni A, Denti MA. Circulating miRNAs as biomarkers for neurodegenerative disorders. Molecules 2014;19:6891–6910.ArticlePubMedPMC
- 44. Kim DH, Yeo SH, Park JM, Choi JY, Lee TH, Park SY, et al. Genetic markers for diagnosis and pathogenesis of Alzheimer’s disease. Gene 2014;545:185–193.ArticlePubMed
- 45. Rebane A, Akdis CA. MicroRNAs in allergy and asthma. Curr Allergy Asthma Rep 2014;14:424.ArticlePubMed
- 46. Rupani H, Sanchez-Elsner T, Howarth P. MicroRNAs and respiratory diseases. Eur Respir J 2013;41:695–705.ArticlePubMed
- 47. D’Alessandra Y, Devanna P, Limana F, Straino S, Di Carlo A, Brambilla PG, et al. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J 2010;31:2765–2773.ArticlePubMedPMCPDF
- 48. Landi MT, Zhao Y, Rotunno M, Koshiol J, Liu H, Bergen AW, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res 2010;16:430–441.ArticlePubMedPMC
- 49. Andorfer CA, Necela BM, Thompson EA, Perez EA. MicroRNA signatures: clinical biomarkers for the diagnosis and treatment of breast cancer. Trends Mol Med 2011;17:313–319.ArticlePubMed
- 50. Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature 2011;473:317–325.ArticlePubMedPDF
- 51. Harraz MM, Dawson TM, Dawson VL. MicroRNAs in Parkinson’s disease. J Chem Neuroanat 2011;42:127–130.ArticlePubMedPMC
- 52. Hwang DY, Ardayfio P, Kang UJ, Semina EV, Kim KS. Selective loss of dopaminergic neurons in the substantia nigra of Pitx3-deficient aphakia mice. Brain Res Mol Brain Res 2003;114:123–131.ArticlePubMed
- 53. Nunes I, Tovmasian LT, Silva RM, Burke RE, Goff SP. Pitx3 is required for development of substantia nigra dopaminergic neurons. Proc Natl Acad Sci U S A 2003;100:4245–4250.ArticlePubMedPMC
- 54. Martinat C, Bacci JJ, Leete T, Kim J, Vanti WB, Newman AH, et al. Cooperative transcription activation by Nurr1 and Pitx3 induces embryonic stem cell maturation to the midbrain dopamine neuron phenotype. Proc Natl Acad Sci U S A 2006;103:2874–2879.ArticlePubMedPMC
- 55. Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science 2007;317:1220–1224.ArticlePubMedPMC
- 56. Miñones-Moyano E, Porta S, Escaramís G, Rabionet R, Iraola S, Kagerbauer B, et al. MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum Mol Genet 2011;20:3067–3078.ArticlePubMed
- 57. Kabaria S, Choi DC, Chaudhuri AD, Mouradian MM, Junn E. Inhibition of miR-34b and miR-34c enhances α-synuclein expression in Parkinson’s disease. FEBS Lett 2015;589:319–325.ArticlePubMedPMC
- 58. Wang G, van der Walt JM, Mayhew G, Li YJ, Züchner S, Scott WK, et al. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am J Hum Genet 2008;82:283–289.ArticlePubMedPMC
- 59. Margis R, Margis R, Rieder CR. Identification of blood microRNAs associated to Parkinsonĭs disease. J Biotechnol 2011;152:96–101.ArticlePubMed
- 60. Cole SL, Vassar R. The role of amyloid precursor protein processing by BACE1, the beta-secretase, in Alzheimer disease pathophysiology. J Biol Chem 2008;283:29621–29625.ArticlePubMedPMC
- 61. Hébert SS, Horré K, Nicolaï L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A 2008;105:6415–6420.ArticlePubMedPMC
- 62. Vassar R, Kovacs DM, Yan R, Wong PC. The beta-secretase enzyme BACE in health and Alzheimer’s disease: regulation, cell biology, function, and therapeutic potential. J Neurosci 2009;29:12787–12794.ArticlePubMedPMC
- 63. Shioya M, Obayashi S, Tabunoki H, Arima K, Saito Y, Ishida T, et al. Aberrant microRNA expression in the brains of neurodegenerative diseases: miR-29a decreased in Alzheimer disease brains targets neurone navigator 3. Neuropathol Appl Neurobiol 2010;36:320–330.ArticlePubMed
- 64. Nelson PT, Kiriakidou M, Mourelatos Z, Tan GS, Jennings MH, Xie K, et al. High-throughput experimental studies to identify miRNA targets directly, with special focus on the mammalian brain. Brain Res 2010;1338:122–130.ArticlePubMedPMC
- 65. Wang WX, Huang Q, Hu Y, Stromberg AJ, Nelson PT. Patterns of microRNA expression in normal and early Alzheimer’s disease human temporal cortex: white matter versus gray matter. Acta Neuropathol 2011;121:193–205.ArticlePubMedPMC
- 66. Goodall EF, Heath PR, Bandmann O, Kirby J, Shaw PJ. Neuronal dark matter: the emerging role of microRNAs in neurodegeneration. Front Cell Neurosci 2013;7:178.ArticlePubMedPMC
- 67. Hébert SS, Papadopoulou AS, Smith P, Galas MC, Planel E, Silahtaroglu AN, et al. Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum Mol Genet 2010;19:3959–3969.ArticlePubMedPDF
- 68. Fenn AM, Smith KM, Lovett-Racke AE, Guerau-de-Arellano M, Whitacre CC, Godbout JP. Increased micro-RNA 29b in the aged brain correlates with the reduction of insulin-like growth factor-1 and fractalkine ligand. Neurobiol Aging 2013;34:2748–2758.ArticlePubMedPMC
- 69. Hooper C, Meimaridou E, Tavassoli M, Melino G, Lovestone S, Killick R. p53 is upregulated in Alzheimer’s disease and induces tau phosphorylation in HEK293a cells. Neurosci Lett 2007;418:34–37.ArticlePubMedPMC
- 70. Zovoilis A, Agbemenyah HY, Agis-Balboa RC, Stilling RM, Edbauer D, Rao P, et al. MicroRNA-34c is a novel target to treat dementias. EMBO J 2011;30:4299–4308.ArticlePubMedPMC
- 71. Kim J, Yoon H, Ramírez CM, Lee SM, Hoe HS, FernándezHernando C, et al. MiR-106b impairs cholesterol efflux and increases Aβ levels by repressing ABCA1 expression. Exp Neurol 2012;235:476–483.ArticlePubMedPMC
- 72. Schonrock N, Humphreys DT, Preiss T, Götz J. Target gene repression mediated by miRNAs miR-181c and miR-9 both of which are down-regulated by amyloid-β. J Mol Neurosci 2012;46:324–335.ArticlePubMed
- 73. Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport 2007;18:297–300.ArticlePubMed
- 74. Lukiw WJ, Zhao Y, Cui JG. An NF-kappaB-sensitive micro RNA-146a-mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. J Biol Chem 2008;283:31315–31322.ArticlePubMedPMC
- 75. Van den Hove DL, Kompotis K, Lardenoije R, Kenis G, Mill J, Steinbusch HW, et al. Epigenetically regulated microRNAs in Alzheimer’s disease. Neurobiol Aging 2014;35:731–745.ArticlePubMed
- 76. Zuccato C, Tartari M, Crotti A, Goffredo D, Valenza M, Conti L, et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet 2003;35:76–83.ArticlePubMedPDF
- 77. Savas JN, Makusky A, Ottosen S, Baillat D, Then F, Krainc D, et al. Huntington’s disease protein contributes to RNA-mediated gene silencing through association with Argonaute and P bodies. Proc Natl Acad Sci U S A 2008;105:10820–10825.ArticlePubMedPMC
- 78. Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J Neurosci 2008;28:14341–14346.ArticlePubMedPMC
- 79. Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, et al. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci U S A 2005;102:16426–16431.ArticlePubMedPMC
- 80. Johnson R, Zuccato C, Belyaev ND, Guest DJ, Cattaneo E, Buckley NJ. A microRNA-based gene dysregulation pathway in Huntington’s disease. Neurobiol Dis 2008;29:438–445.ArticlePubMed
- 81. Jin R. Quantum sized, thiolate-protected gold nanoclusters. Nanoscale 2010;2:343–362.ArticlePubMed
- 82. Schmid G. Large clusters and colloids. Metals in the embryonic state. Chem Rev 1992;92:1709–1727.Article
- 83. Kubo R. Electronic properties of metallic fine particles. I. J Phys Soc Jpn 1962;17:975–986.Article
- 84. Dupree R, Smithard MA. The electronic properties of small metal particles: the electric polarizability. J Phys C 1972;5:408.Article
- 85. Choi S, Dickson RM, Yu J. Developing luminescent silver nanodots for biological applications. Chem Soc Rev 2012;41:1867–1891.ArticlePubMed
- 86. Latorre A, Somoza Á. DNA-mediated silver nanoclusters: synthesis, properties and applications. Chembiochem 2012;13:951–958.ArticlePubMed
- 87. Obliosca JM, Liu C, Batson RA, Babin MC, Werner JH, Yeh HC. DNA/RNA detection using DNA-templated few-atom silver nanoclusters. Biosensors (Basel) 2013;3:185–200.ArticlePubMedPMC
- 88. Le Guével X. Recent advances on the synthesis of metal quantum nanoclusters and their application for bioimaging. IEEE J Sel Top Quantum Electron 2014;20:12.
- 89. Patel SA, Richards CI, Hsiang JC, Dickson RM. Water-soluble Ag nanoclusters exhibit strong two-photon-induced fluorescence. J Am Chem Soc 2008;130:11602–11603.ArticlePubMedPMC
- 90. Richards CI, Choi S, Hsiang JC, Antoku Y, Vosch T, Bongiorno A, et al. Oligonucleotide-stabilized Ag nanocluster fluorophores. J Am Chem Soc 2008;130:5038–5039.ArticlePubMedPMC
- 91. Petty JT, Giri B, Miller IC, Nicholson DA, Sergev OO, Banks TM, et al. Silver clusters as both chromophoric reporters and DNA ligands. Anal Chem 2013;85:2183–2190.ArticlePubMedPMC
- 92. Shah P, Thulstrup PW, Cho SK, Bjerrum MJ, Yang SW. DNA-RNA chimera indicates the flexibility of the backbone influences the encapsulation of fluorescent AgNC emitters. Chem Commun (Camb) 2014;50:13592–13595.ArticlePubMed
- 93. Shah P, Choi SW, Kim HJ, Cho SK, Thulstrup PW, Bjerrum MJ, et al. DNA/RNA chimera templates improve the emission intensity and target the accessibility of silver nanocluster-based sensors for human microRNA detection. Analyst 2015;140:3422–3430.ArticlePubMed
- 94. Shah P, Choi SW, Kim HJ, Cho SK, Bhang YJ, Ryu MY, et al. Locking-to-unlocking system is an efficient strategy to design DNA/silver nanoclusters (AgNCs) probe for human miRNAs. Nucleic Acids Res 2016;44:e57.ArticlePubMedPMCPDF
- 95. Cho SK, Ben Chaabane S, Shah P, Poulsen CP, Yang SW. COP1 E3 ligase protects HYL1 to retain microRNA biogenesis. Nat Commun 2014;5:5867.ArticlePubMedPDF
- 96. Zhang M, Liu YQ, Yu CY, Yin BC, Ye BC. Multiplexed detection of microRNAs by tuning DNA-scaffolded silver nanoclusters. Analyst 2013;138:4812–4817.ArticlePubMed
- 97. Xia X, Hao Y, Hu S, Wang J. Hairpin DNA probe with 5’-TCC/CCC-3’ overhangs for the creation of silver nanoclusters and miRNA assay. Biosens Bioelectron 2014;51:36–39.ArticlePubMed
- 98. Dong H, Hao K, Tian Y, Jin S, Lu H, Zhou SF, et al. Label-free and ultrasensitive microRNA detection based on novel molecular beacon binding readout and target recycling amplification. Biosens Bioelectron 2014;53:377–383.ArticlePubMed
- 99. Koshiol J, Wang E, Zhao Y, Marincola F, Landi MT. Strengths and limitations of laboratory procedures for microRNA detection. Cancer Epidemiol Biomarkers Prev 2010;19:907–911.ArticlePubMedPMC
REFERENCES
Figure & Data
References
Citations

- Nanosensors for the diagnosis and therapy of neurodegenerative disorders and inflammatory bowel disease
Thirunavukkarsu Palaniyandi, Kanagavalli B, Pranav Prabhakaran, Sandhiya Viswanathan, Mugip Rahaman Abdul Wahab, Sudhakar Natarajan, Senthil Kumar Kaliya Moorthy, Saravanan Kumarasamy
Acta Histochemica.2023; 125(2): 151997. CrossRef - Emerging role of microRNAs and long non-coding RNAs in COVID-19 with implications to therapeutics
Kaifee Arman, Zeinab Dalloul, Esra Bozgeyik
Gene.2023; 861: 147232. CrossRef - Biofluid Biomarkers in the Prognosis of Amyotrophic Lateral Sclerosis: Recent Developments and Therapeutic Applications
Daniel Sanchez-Tejerina, Arnau Llaurado, Javier Sotoca, Veronica Lopez-Diego, Jose M. Vidal Taboada, Maria Salvado, Raul Juntas-Morales
Cells.2023; 12(8): 1180. CrossRef - Rapid detection of SARS-CoV-2 genetic targets using nanoporous waveguide based competitive displacement assay
Megan Makela, Zhihai Lin, Pao Tai Lin
Giant.2023; 15: 100173. CrossRef - Micro Nanoengraving Technology and Aesthetic Practice of Architectural Sculpture Art
Weili Zhu, Dong Wei, Awais Ahmed
Journal of Nanomaterials.2022; 2022: 1. CrossRef - Morphine-mediated release of astrocyte-derived extracellular vesicle miR-23a induces loss of pericyte coverage at the blood-brain barrier: Implications for neuroinflammation
Ke Liao, Fang Niu, Guoku Hu, Shilpa Buch
Frontiers in Cell and Developmental Biology.2022;[Epub] CrossRef - A Review on the Role of Nanosensors in Detecting Cellular miRNA Expression in Colorectal Cancer
Koyeli Girigoswami, Agnishwar Girigoswami
Endocrine, Metabolic & Immune Disorders - Drug Targets.2021; 21(1): 12. CrossRef - Duplex DNA-functionalized graphene oxide: A versatile platform for miRNA sensing
Bomi Shin, Woo-Keun Kim, Seokjoo Yoon, Jieon Lee
Sensors and Actuators B: Chemical.2020; 305: 127471. CrossRef - Recent advances in the bioanalytical and biomedical applications of DNA-templated silver nanoclusters
Jiaqi Xu, Xuanmeng Zhu, Xi Zhou, Farjana Yeasmin Khusbu, Changbei Ma
TrAC Trends in Analytical Chemistry.2020; 124: 115786. CrossRef - Identification of Ovine Serum miRNAs Following Bacterial Lipopolysaccharide Challenge
Ankita Sharma, Umesh K. Shandilya, Tianna Sullivan, Danielle Naylor, Angela Canovas, Bonnie A. Mallard, Niel A. Karrow
International Journal of Molecular Sciences.2020; 21(21): 7920. CrossRef - Epigenetics in amyotrophic lateral sclerosis: a role for histone post-translational modifications in neurodegenerative disease
Seth A. Bennett, Royena Tanaz, Samantha N. Cobos, Mariana P. Torrente
Translational Research.2019; 204: 19. CrossRef - Network-based approach to identify molecular signatures and therapeutic agents in Alzheimer’s disease
Md. Rezanur Rahman, Tania Islam, Beste Turanli, Toyfiquz Zaman, Hossain Md. Faruquee, Md. Mafizur Rahman, Md. Nurul Haque Mollah, Ranjan Kumar Nanda, Kazim Yalcin Arga, Esra Gov, Mohammad Ali Moni
Computational Biology and Chemistry.2019; 78: 431. CrossRef - Generic Neutravidin Biosensor for Simultaneous Multiplex Detection of MicroRNAs via Electrochemically Encoded Responsive Nanolabels
Sawsen Azzouzi, Zina Fredj, Anthony P. F. Turner, Mounir Ben Ali, Wing Cheung Mak
ACS Sensors.2019; 4(2): 326. CrossRef - MiR‐140 modulates the inflammatory responses of Mycobacterium tuberculosis‐infected macrophages by targeting TRAF6
Xiaofei Li, Shan Huang, Tingting Yu, Guiliang Liang, Hongwei Liu, Dong Pu, Niancai Peng
Journal of Cellular and Molecular Medicine.2019; 23(8): 5642. CrossRef - Mass spectrometry: A platform for biomarker discovery and validation for Alzheimer's and Parkinson's diseases
Eugene M. Cilento, Lorrain Jin, Tessandra Stewart, Min Shi, Lifu Sheng, Jing Zhang
Journal of Neurochemistry.2019; 151(4): 397. CrossRef - MicroRNA Assisted Gene Regulation in Colorectal Cancer
Adewale Fadaka, Ashley Pretorius, Ashwil Klein
International Journal of Molecular Sciences.2019; 20(19): 4899. CrossRef - Circulating miRNAs, Small but Promising Biomarkers for Autism Spectrum Disorder
Salam Salloum-Asfar, Noothan J. Satheesh, Sara A. Abdulla
Frontiers in Molecular Neuroscience.2019;[Epub] CrossRef - A highly sensitive miR-195 nanobiosensor for early detection of Parkinson’s disease
Zahra Aghili, Navid Nasirizadeh, Adeleh Divsalar, Shahram Shoeibi, Parichehreh Yaghmaei
Artificial Cells, Nanomedicine, and Biotechnology.2018; 46(sup1): 32. CrossRef - Quantitative and multiplex microRNA assays from unprocessed cells in isolated nanoliter well arrays
Augusto M. Tentori, Maxwell B. Nagarajan, Jae Jung Kim, Wen Cai Zhang, Frank J. Slack, Patrick S. Doyle
Lab on a Chip.2018; 18(16): 2410. CrossRef - The Role of MicroRNAs in Patients with Amyotrophic Lateral Sclerosis
Efthimios Dardiotis, Athina-Maria Aloizou, Vasileios Siokas, George P. Patrinos, Georgia Deretzi, Panayiotis Mitsias, Michael Aschner, Aristidis Tsatsakis
Journal of Molecular Neuroscience.2018; 66(4): 617. CrossRef
Comments on this article
 KMDS
KMDS
 E-submission
E-submission
 PubReader
PubReader ePub Link
ePub Link Cite
Cite